This study identified a mutated form of SARS-CoV-2 called Spike D614G that became very common in March 2020, but this study does not show that this mutation actually makes the virus more contagious or deadly.
Key takeaways
- Spike D614G became the dominant form of SARS-CoV-2 during March 2020.
- Spike D614G may have made the virus more contagious, but it’s also possible that Spike D614G became more common due to random chance. For example, it may have arisen from a founder effect in the European population that later spread to other parts of the world.
- In clinical data from more than 400 patients, the new D614G form was not statistically significantly more contagious or lethal than the prior form.
- Further studies are needed to determine the exact role of Spike D614G in the transmissibility and lethality of SARS-CoV-2.
Why is this important?
Headlines in early May 2020 touted claims of a new SARS-CoV-2 strain (Spike D614G) that is more contagious and possibly more deadly. In addition to potentially worsening the pandemic, a new mutation could potentially impact efforts to develop vaccines or drugs against the virus.
What is Spike D614G?
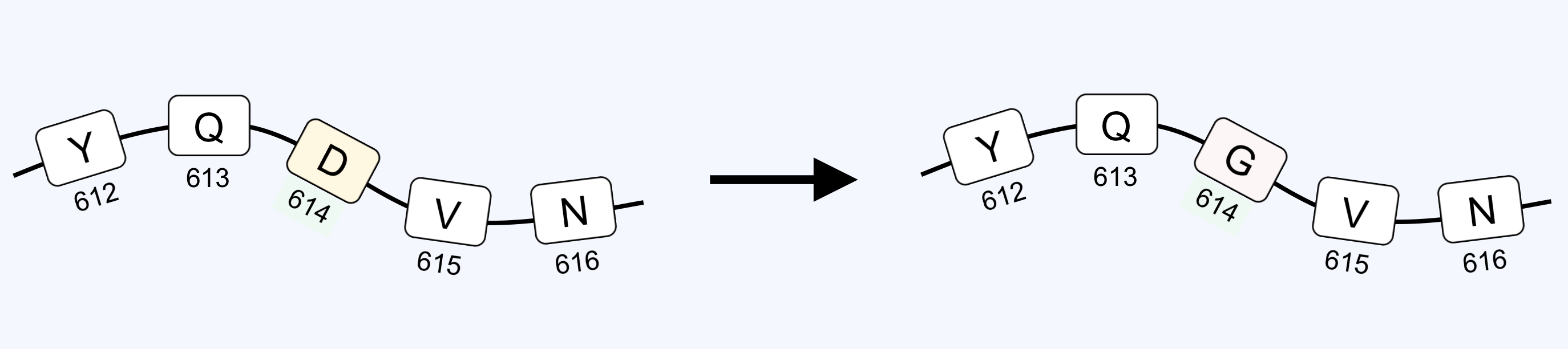
The protein in the SARS-CoV-2 coronavirus that is responsible for recognizing and entering human cells in the lung is called the spike protein. This protein is composed of 1,276 amino acids, and is the target for almost every vaccine currently in development.
Amino acid ‘aspartic acid’ (D) was found at position 614 in the initial sequences obtained for this virus. This study, however, has shown that in many of the sequences from more recent patients, amino acid ‘glycine’ (G) can be found in this position. Therefore, the mutation is called D614G, meaning amino acid D at position 614 was mutated to G. There are now two main forms of the virus: the initial form with amino acid D in position 614 (D614), and the new mutated form with amino acid G at this position (G614).
What did the study do?
-
Sampling
Collected global SARS-CoV-2 sequence data from the GISAID database
Our Take
The researchers used SARS-CoV-2 genome sequences from the GISAID database. They built an automated pipeline to download and clean the sequence data before analyzing it.
GISAID database contains high-quality sequence data on SARS-CoV-2
The authors employed sequence data from the Global Initiative for Sharing All Influenza Data (GISAID) database, a high-quality repository of SARS-CoV-2 sequences from around the world.
GISAID database has a small number of recent samples
The study had only 7 viral sequences from the time period March 31 – April 6 in North America. However, the study did include more than 100 sequences from this time period in Europe.
-
Analysis
Analyzed the prevalence of mutations at 14 different sites in SARS-CoV-2 genomes over time
The researchers identified 14 different locations in the SARS-CoV-2 genome that contained mutations, and they studied how these mutations changed over time. They found that Spike D614G became the most common strain of SARS-CoV-2 during March 2020.
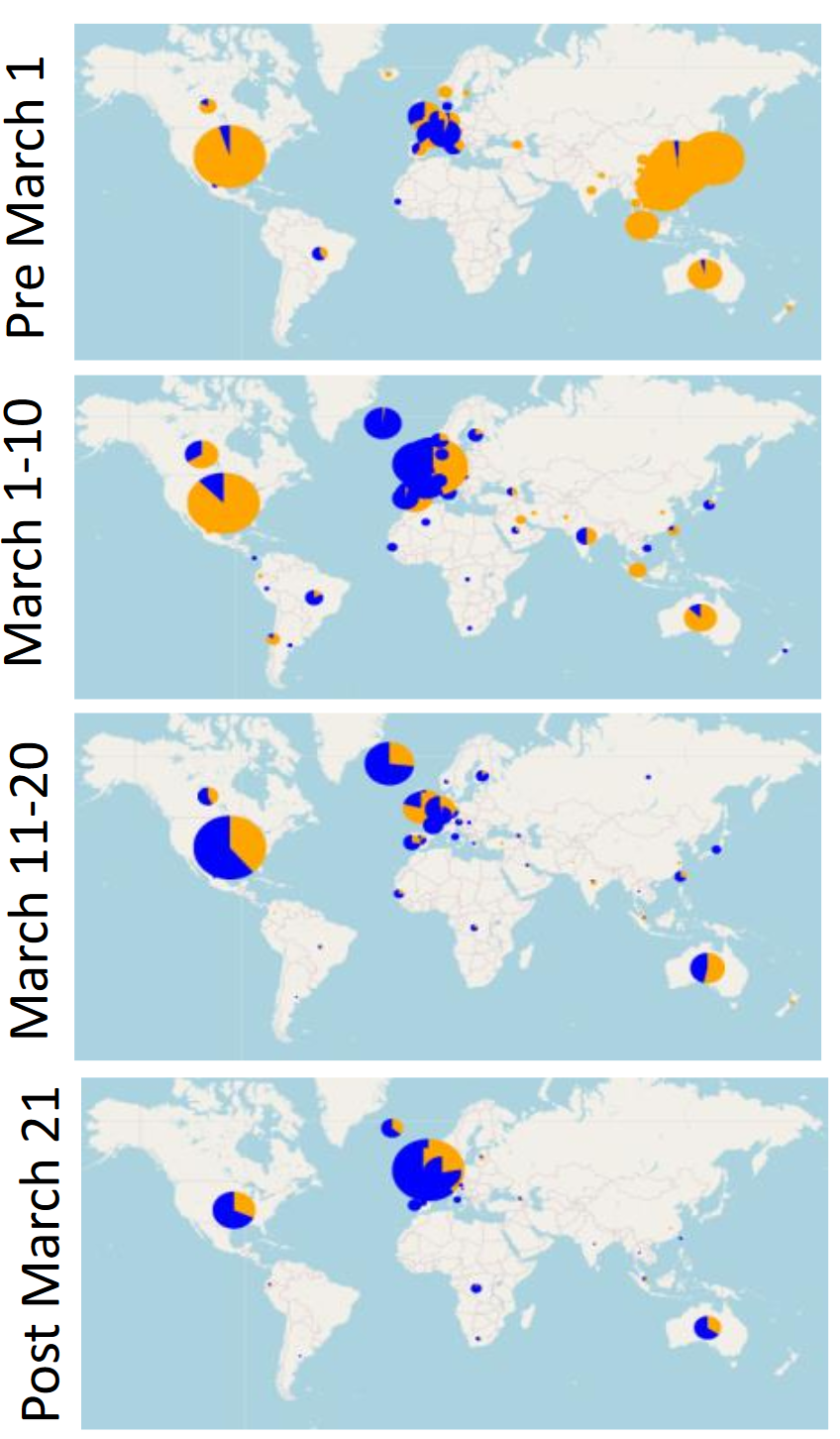
Blue represents Spike D614G
Our Take
Sensible analysis choices
The authors identified 14 different sites within the SARS-CoV-2 genome that had a mutation with at least 0.3% prevalence. The authors further chose to focus on mutations in the Spike protein, because the Spike protein is important for viral entry into the host cell.
Convincing primary result
The authors found that a large number of patients had a mutation at amino acid position 614 in the Spike protein. They demonstrated that this mutation, called Spike D614G, became the most common form of SARS-CoV-2 during March 2020.
Poorly supported speculation
The researchers speculated that the Spike D614G mutation might have become more common by helping the SARS-CoV-2 virus enter human cells more efficiently, or by helping it to evade the immune system. However, they did not provide any experimental evidence to support these claims.
-
Impact
Analyzed clinical outcomes of more than 400 patients
Our Take
Not enough information in study of clinical outcomes
The researchers analyzed a database of individuals in Sheffield, UK, to see if patients with either the original form or the D614G mutation differed in their risk of being admitted to the hospital or the ICU. The researchers did not find a statistically significant difference in the clinical outcomes between the two forms of the virus. The researchers did show a small increase in viral load for patients with the D614G form of the virus. No information was provided about comorbidities for the different cohorts. However, comorbidities have a large impact on SARS-CoV-2 outcomes 1 2, so including this information is essential for interpreting the results. Also, the number of patients included was not consistent throughout the paper. In the text of the paper, the authors wrote that the clinical study included 453 patients. In Figure 5 of the paper, they wrote that the clinical study included 447 patients. In Figure 5C, they included data from only 433 patients. The discrepancies in these numbers were not clearly explained.
-
Conclusion
Spike D614G form of SARS-CoV-2 is more transmissible than previous strains
Our Take
Limited support for conclusion
The authors speculated that Spike D614G is likely to be more transmissible than the previous strain (Spike D614) due to its rising prevalence over time. The authors speculated about the possible role of the Spike D614G mutation in enhancing viral binding to the host receptor or evading the immune system. However, they did not perform experiments to support these speculations.
Furthermore, the researchers did not adequately account for founder effects in the work. While the dominant form of the virus in Wuhan was the original variant, even early on in parts of Europe the dominant form was D614G. This could be consistent with a founder effect, where the initial forms of the virus brought to Europe carried this mutation. Soon after, travel was restricted to China by much of the world, and those who had come from Wuhan were identified and quarantined, however travel within Europe as well as international travel out of Europe was not restricted, and many of those passengers did not undergo quarantine procedures until much later. It is possible that the restrictions placed on travelers originating from China and the lack of restrictions on European passengers could have allowed the D614G variant of the virus to be spread throughout the world and initiate new viral colonies, making it more common than the original variant that initially arrived from Wuhan.